Salami Server
Project description:
Multiple Structure Alignment
Salami Server at Center for Bioinformatics Hamburg
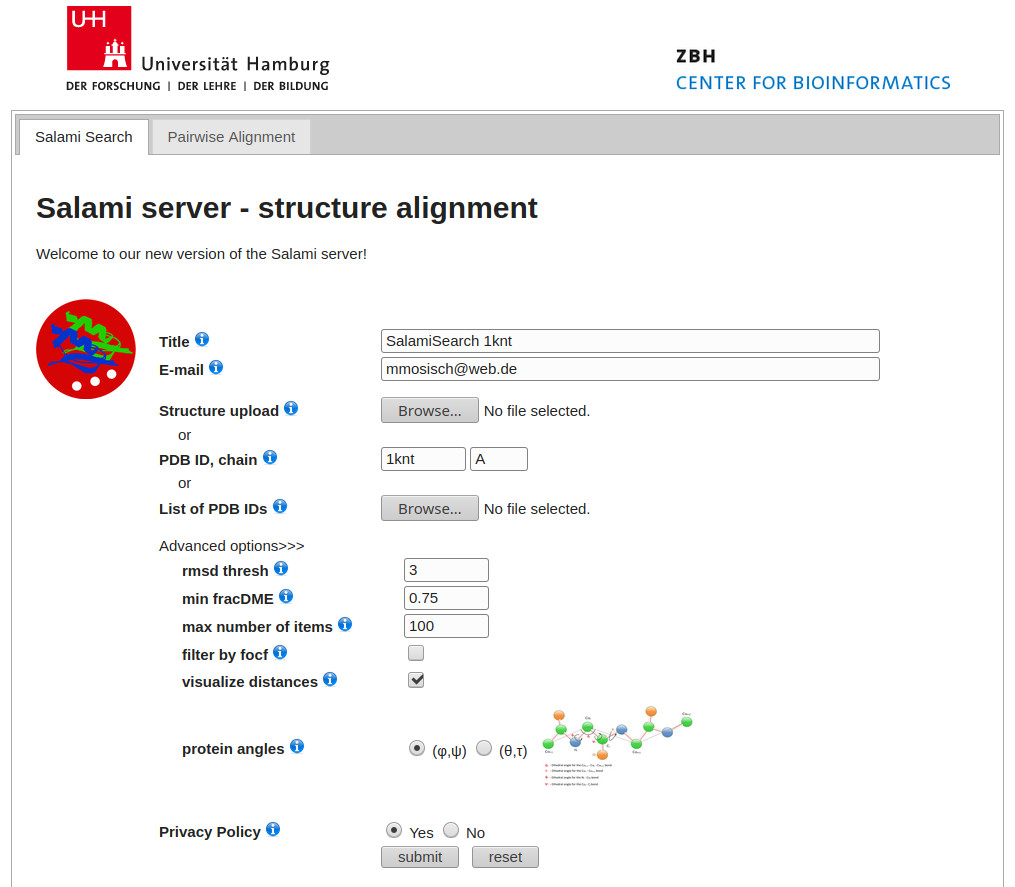
Upper Image : Query Form
Given an input structure (pdb upload file, pdb id or pdb list) one is able to find within representative structures for the particular PDB clusters (currently 53.000) the 100 most similar structures without any sequence information. The calculations are quite fast and can be efficiently performed on the workstations of the local student pool cluster in the student's excersise room. An ordinary query takes usually about 10 minutes but can take up to 60 minutes. Thus you are informed immidiately via Email as soon as the comparison calculations have finished.
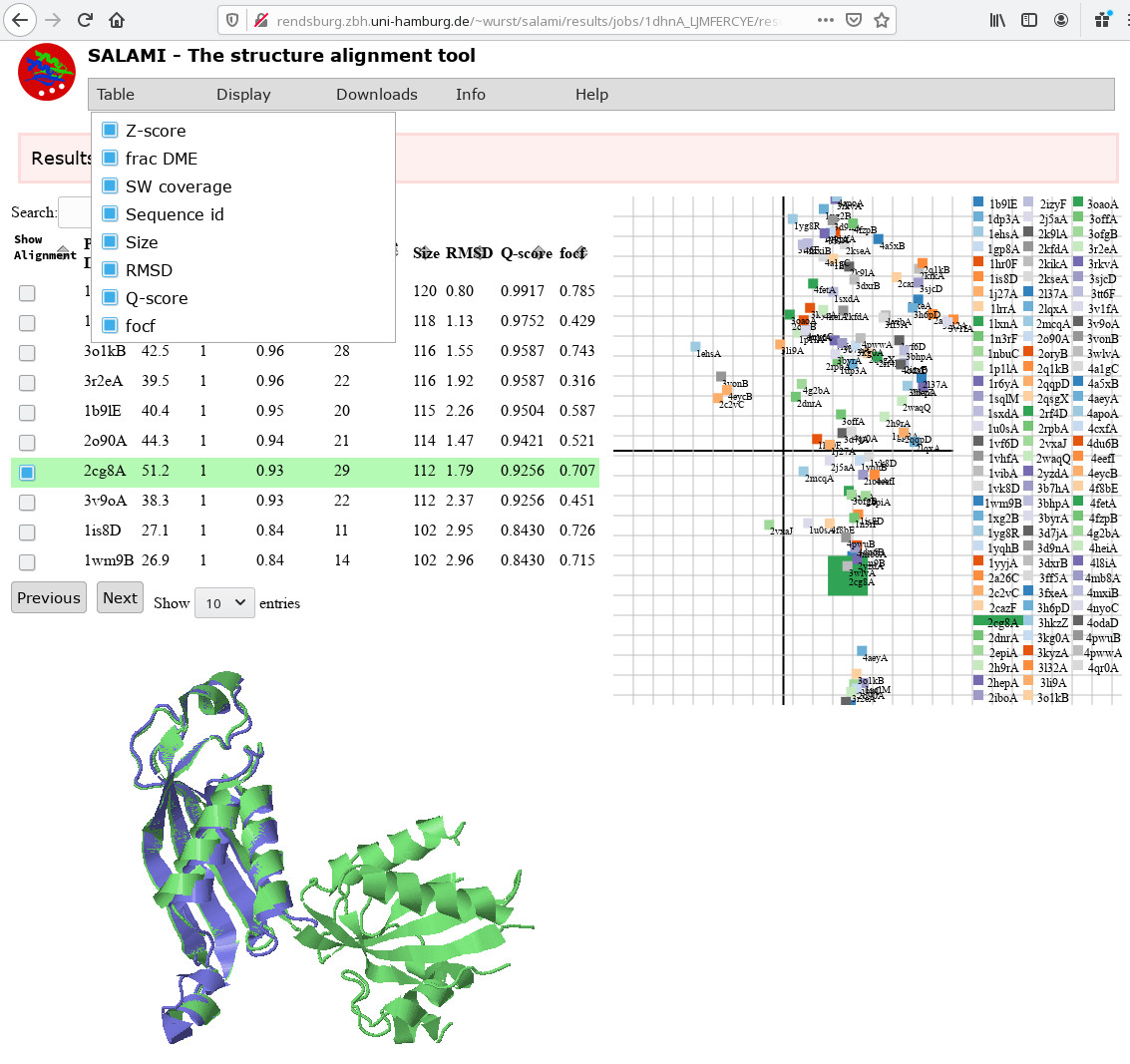
Lower image : Query Result
The result are the first 100 most similar structures that can be found in the protein database and their illustration. Pairwise the structures are visual inspectable in the current Jmol (Jsmol) protein structure viewer in form of javascript applet. You can also investigate a couple of key performance indices amongst others RMSD, fracDME, and Z-score. Further more the result space has been reduced to two dimensions, so one can have a look how the 100 most similar structures w.r.t. the input structure are spread in space.
Since the project was broken and taken offline for at least 3 years, there was the need for a resurrection. Now we are lucky to announce that I have brought the Salami Server back to life.
Computational skills: Clustering, Principle Component Analysis, Sun Grid Engine, AnsiC, Perl, PHP, Javascript and HTML. cite ref
Multiple Structure Alignment
Salami Server at Center for Bioinformatics Hamburg
Upper Image : Query Form
Given an input structure (pdb upload file, pdb id or pdb list) one is able to find within representative structures for the particular PDB clusters (currently 53.000) the 100 most similar structures without any sequence information. The calculations are quite fast and can be efficiently performed on the workstations of the local student pool cluster in the student's excersise room. An ordinary query takes usually about 10 minutes but can take up to 60 minutes. Thus you are informed immidiately via Email as soon as the comparison calculations have finished.
Lower image : Query Result
The result are the first 100 most similar structures that can be found in the protein database and their illustration. Pairwise the structures are visual inspectable in the current Jmol (Jsmol) protein structure viewer in form of javascript applet. You can also investigate a couple of key performance indices amongst others RMSD, fracDME, and Z-score. Further more the result space has been reduced to two dimensions, so one can have a look how the 100 most similar structures w.r.t. the input structure are spread in space.
Since the project was broken and taken offline for at least 3 years, there was the need for a resurrection. Now we are lucky to announce that I have brought the Salami Server back to life.
Computational skills: Clustering, Principle Component Analysis, Sun Grid Engine, AnsiC, Perl, PHP, Javascript and HTML. cite ref

Salami Server
Friday, 06. Jun 2019